Data exploration
Here we show how you to manipulate and visualize the incidence and climatic data that we previously created. Before starting, let’s make sure that the required packages are installed:
to_install <- setdiff(c("dplyr", "magrittr", "readr", "sf"), rownames(installed.packages()))
if (length(to_install) > 0) install.packages(to_install)Loading the require packages for direct interaction:
library(magrittr) # for the pipe operator
library(sf) # for the geographic objects methodsIncidence data
Let’s get the dengue data:
dengue <- readr::read_csv("https://raw.githubusercontent.com/choisy/DMo2019/master/data/dengue.csv")You can split the data frame of incidences into a list of data frames, with one data frame per state as so:
dengue_by_state <- split(dengue, dengue$state)From which list you can extract whatever state you want:
dengue_data <- dengue_by_state$TerengganuLet’s define some coordinates to customize the \(x\)-axis:
ats <- dengue_data %>%
dplyr::group_by(year) %>%
dplyr::summarise(at = mean(week)) %>%
dplyr::ungroup() %$%
setNames(at, year) and
first_weeks <- which(dengue_data$week == 1) - .5The following function plots a chronologically-order vector of incidence from the list dengue_by_state:
plot_incidence <- function(x, ...) {
barplot(x, space = 0, axes = FALSE, xlab = NA, ylab = "incidence", ...)
axis(1, first_weeks, FALSE)
axis(1, ats + first_weeks, names(ats), tick = FALSE)
axis(2)
abline(v = first_weeks, lty = 2, col = "grey")
}Let’s use this function to plot the time series of incidence for all the states:
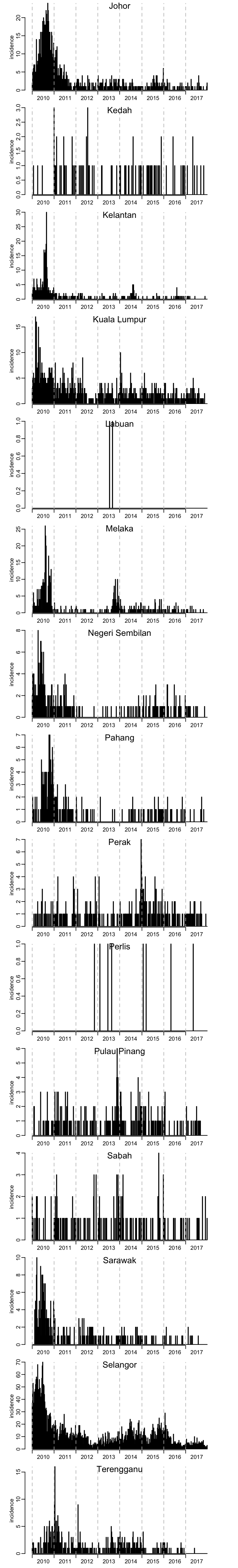
par(mfrow = c(15, 1))
for (state in unique(dengue$state)) {
plot_incidence(dengue_by_state[[state]]$incidence)
mtext(state, line = -1)
}
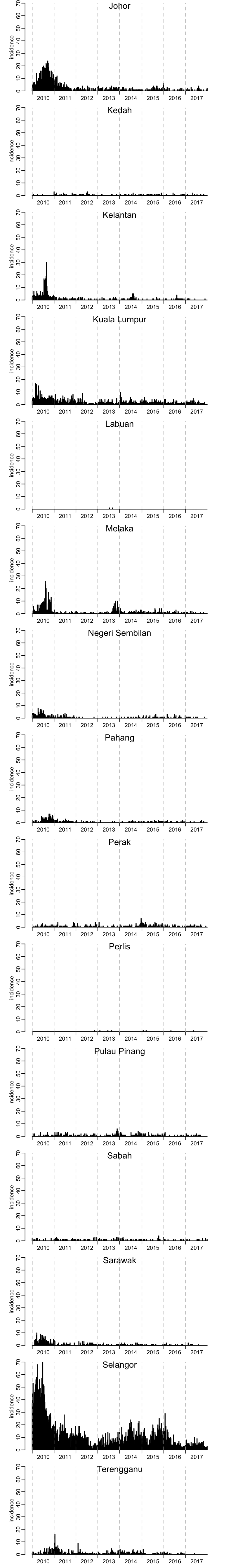
The same, but this time with the same range of values on the \(y\)-axis:
par(mfrow = c(15, 1))
for (state in unique(dengue$state)) {
plot_incidence(dengue_by_state[[state]]$incidence, ylim = c(0, max(dengue$incidence)))
mtext(state, line = -1)
}
Geographic data
We can download maps of any country directly from the GADM website:
malaysia <- readRDS(url("https://biogeo.ucdavis.edu/data/gadm3.6/Rsf/gadm36_MYS_1_sf.rds"))Climatic data
Downloading the climatic stations:
stations <- readr::read_csv("https://raw.githubusercontent.com/choisy/DMo2019/master/data/climatic%20stations.csv")Plotting the locations of the climatic stations. First we need to transform the stations as a geographic object:
stations_geo <- sf::st_as_sf(stations,
coords = c("longitude", "latitude"),
crs = "+proj=longlat +datum=WGS84 +no_defs +ellps=WGS84 +towgs84=0,0,0")malaysia %>%
sf::st_simplify(TRUE, .05) %>%
sf::st_geometry() %>%
plot(col = "lightgrey")
plot(stations_geo, add = TRUE, pch = 3, col = "red")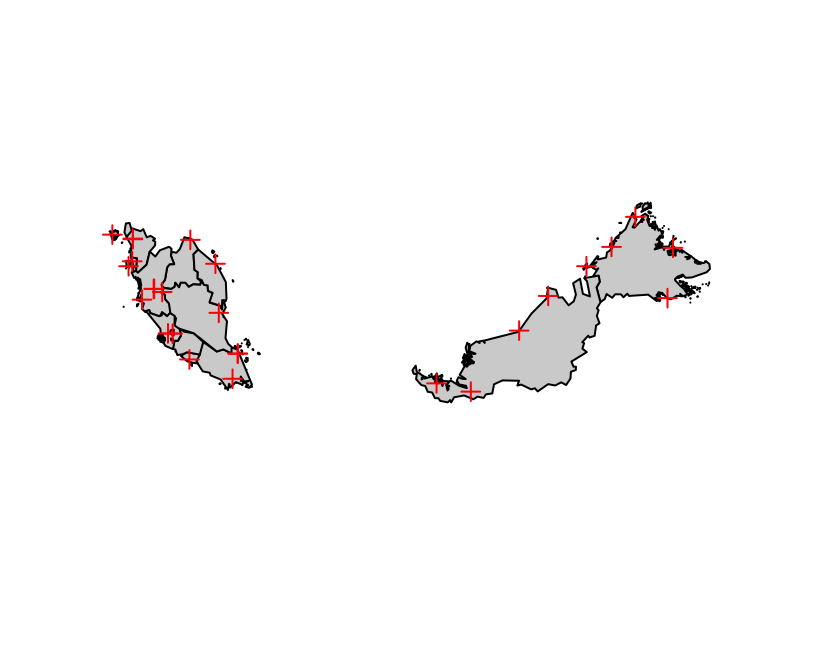
Let’s now download the climatic data:
climate <- readr::read_csv("https://raw.githubusercontent.com/choisy/DMo2019/master/data/climate.csv")Let’s now aggregate these data by epi week so that we can compare more easily with the incidence data that are also defined by epi week:
climate_week <- climate %>%
dplyr::mutate(year = lubridate::year(day),
week = lubridate::epiweek(day)) %>%
dplyr::select(-day) %>%
dplyr::group_by(station, year, week) %>%
dplyr::summarise_all(mean, na.rm = TRUE) %>%
dplyr::ungroup() %>%
dplyr::mutate_at(dplyr::vars(ra, sn, ts, fg), list(~ . > 0)) %>%
dplyr::mutate_all(. %>% ifelse(is.nan(.), NA, .))Relating climatic data to incidence data
Let’s say that we want to focus on the state of Selangor and we want to identify the climatic stations that are in this state:
selangor <- dplyr::filter(malaysia, NAME_1 == "Selangor")
selangor_station <- stations_geo[as.integer(sf::st_contains(selangor, stations_geo)), ]Let’s see that on the map:
plot(sf::st_geometry(selangor), col = "lightgrey")
plot(stations_geo, add = TRUE, pch = 3, col = "red")
plot(selangor_station, add = TRUE, col = "blue")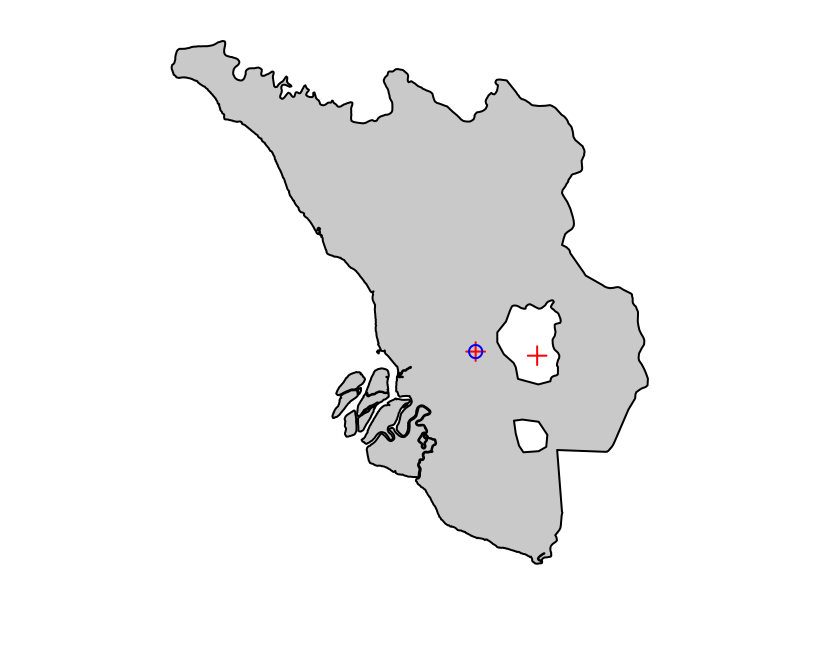
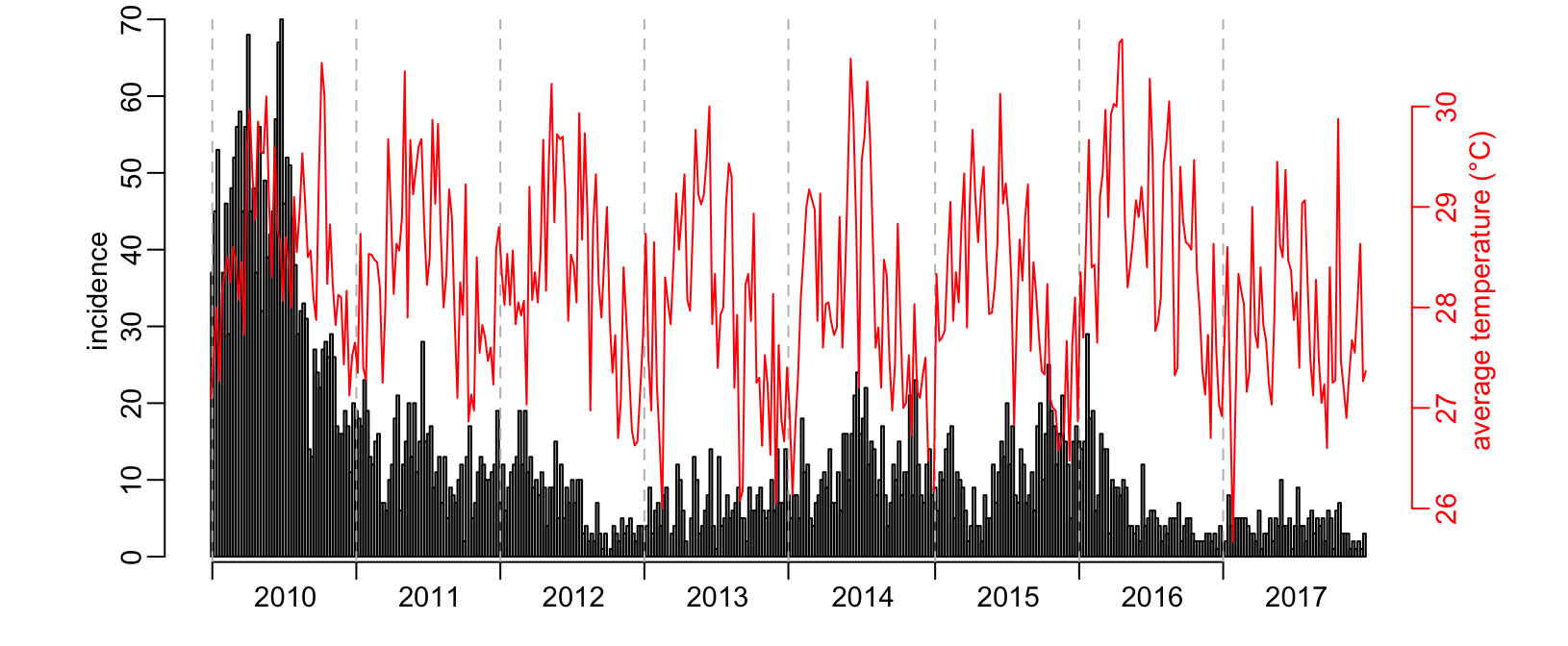
Now we can compare incidence with for example average temperature for the state of Selangor:
plot_incidence(dengue_by_state$Selangor$incidence)
par(new = TRUE)
climate_week %>%
dplyr::filter(station == selangor_station$station) %>%
dplyr::arrange(year, week) %$%
plot(ta, type = "l", col = "red", axes = FALSE, ann = FALSE)
axis(4, col = "red", col.axis = "red")
mtext("average temperature (°C)", 4, 1.5, col = "red")